Single-cell omics platform develops and provides open-access single-cell and spatial transcriptomics services nationally and internationally. Sc omics collaborates very closely with the Biocenter Finland Genome-wide methods and Bioinformatics platforms.
Sc omics platform aims at bringing the latest single cell omics tools and technologies available for the research community. There are services available for single cell genomics (based on Missionbio Tapestri), single cell transcriptomics (Chromium by 10XGenomics), single cell epigenomics (Chromium by 10XGenomics) and single cell proteomics (Helios™ mass cytometry) to enable cutting-edge research in many areas of molecular biology and molecular medicine. In addition to single cell omics our platform has focused on providing services for spatial omics both on cellular level (Visium by 10XGenomics, GeoMX DSP by NanoString and Hyperion by Standard Biotools) and sub-cellular level (spring 2024).
Besides providing services sc omics platform is actively implementing new technologies, developing services, and participating in training of the users.Nodes
UEF: University of Eastern Finland; UH: University of Helsinki; UO: University of Oulu; UTU: University of Turku; ÅAU: Åbo Akademi University.
Contact details
Platform Chair
Tapio Lönnberg
taplon@utu.fi
Platform Vice Chair
Minna Kaikkonen-Määttä
minna.kaikkonen@uef.fi
Services
| Single-cell services provided by sc -omics platform | UH | UH | UEF | UTU | UTU | |
|---|---|---|---|---|---|---|
| Service category | Service | SCA | ScProtGen | SCGC | SCORE | MCy |
| SC transcriptomics (10X Chromium) | 3´gene expression | X | X | X | X | |
| 5´and V(D)J gene expression | X | X | X | |||
| Fixed RNA profiling | X | X | X | |||
| SC transcriptomics (BD Rhapsody) | 3´and V(D)J gene expression | X | ||||
| SC epigenomics (10X Chromium) | SC ATAC-seq | X | X | X | X | |
| SC multiome (Chromium) | ATAC + 3´gene expression | X | X | X | ||
| SC functional genomics | SC CRISPR screens | X | X | X | ||
| SC iso-seq (Kinnex, MAS-seq) | Long read sequencing of cDNA produced by 10XChromium 3´sequencing protocol | X | ||||
| SC genomics (Tapestri) | SC DNA seq | X | ||||
| SC dispensing (cellenONE) | Gentle, image based dispensing of single cells to any kind of plate | X | ||||
| Spatial gene expression (10X Visium) | For FFPE samples | Training | X | |||
| For fresh frozen samples | X | Training | X | |||
| Spatial gene expression (NanoString GeoMX) | Training for customers to use by themselves | X | ||||
| Operator assisted service | ||||||
| Spatial gene expression (subcellular level) | Training for customers to use by themselves | TBA | ||||
| Operator assisted service | TBA | |||||
| Imaging mass cytometry | Operator assisted service | X | ||||
| Imaging mass cytometry | Training for customers to use by themselves | X | ||||
| Mass cytometry | Training for customers to use by themselves | X | ||||
| Mass cytometry | Operator assisted service | X | ||||
Recent user publications
Karaman et al. 2022, Interplay of vascular endothelial growth factor receptors in organ-specific vessel maintenance. J Exp Med (2022) 219 (3): e20210565.https://doi.org/10.1084/jem.20210565
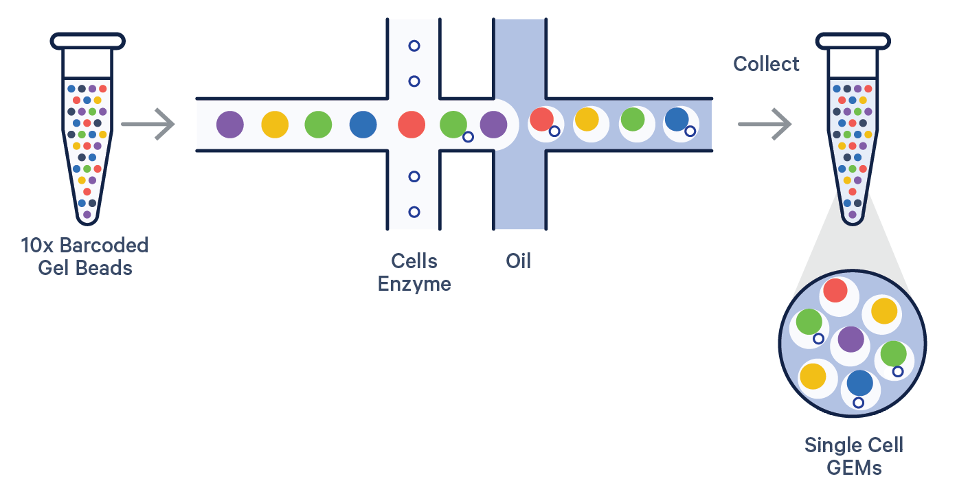
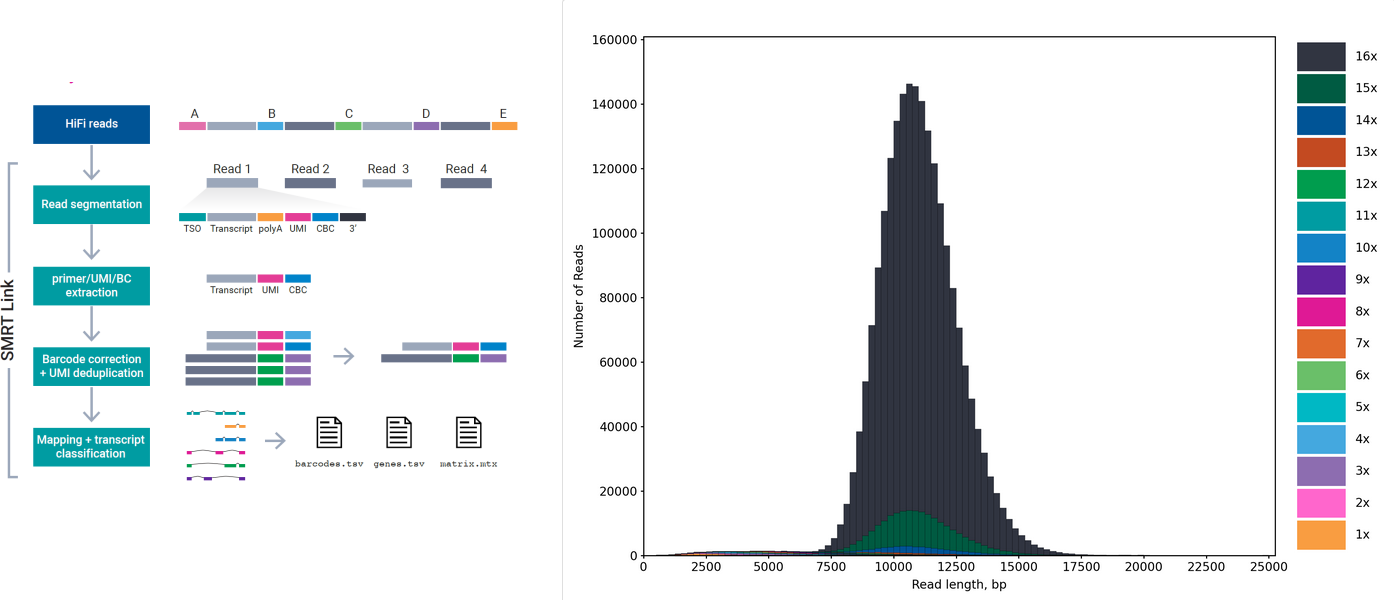
Schematic figure caption of the MAS-seq protocol (left) and size distribution of a MAS-seq data produced from a 10x single cell cDNA library with PacBio Sequel 2 machine (right).


